library(dplyr)
library(magrittr)
library(ggplot2)
library(reactable)
data("mtcars")
reactable(
mtcars,
defaultPageSize = 5,
columns = list(
.rownames = colDef(
name = "model",
sortable = TRUE,
minWidth = 200)
)
)Introduction
Correlations are one of the simplest ways to look at the association between 2 variables. However, I often find myself running correlations on many variables when exploring a new data set. I was tired of writing cor(x, y) and I wanted a quick way to visualize correlations, so I created a correlation matrix using ggplot2.
Data
Let’s explore the mtcars data set and create a correlation matrix.
In the mtcars data set above, there are 11 columns and 32 rows of data.
The columns include: mpg, cyl, disp, hp, drat, wt, qsec, vs, am, gear, carb.
Here is a description of all the variables.
| Variable | Description |
|---|---|
| mpg | Mile per gallon |
| cyl | Number of Cylinders |
| disp | Displacement |
| hp | Gross Horsepower |
| drat | Rear Axle Ratio |
| wt | Weight (lb/1000) |
| qsec | Quarter Mile Time |
| vs | Straight Engine |
| am | Manual Transmission |
| gear | Number of Forward Gears |
| carb | Number of Carburetors |
The Final Product
This is the correlation matrix that we will be creating and I’ve added the code to a function at the end of this post. For those interested in learning, I’ve provided a description of the key steps in getting the correlations and processing the data before creating the correlation matrix. Otherwise, feel free to jump ahead to the bottom of the post to see a link to the cor_matrix function.
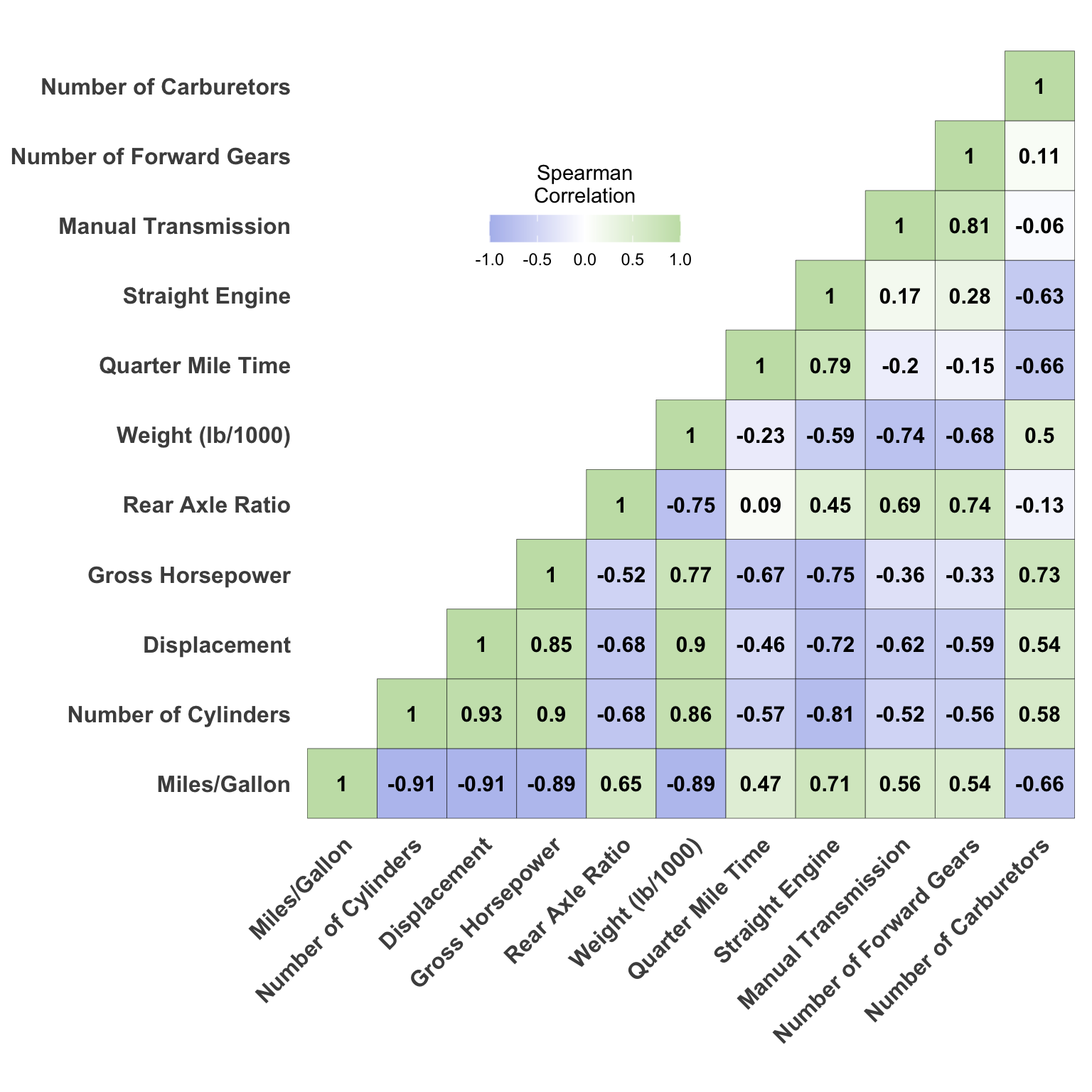
Correlations
To create the correlation matrix, we will use the rcorr function from the Hmisc package. First, we need to take the mtcars data set and convert it to a matrix before calling the Hmisc::rcorr function. We will use spearman correlations for this example, but the type argument can also be switched to pearson.
vars <- c("mpg", "cyl", "disp", "hp", "drat",
"wt", "qsec", "vs", "am", "gear", "carb")
corres <-
mtcars %>%
select(all_of(vars)) %>%
as.matrix() %>%
Hmisc::rcorr(., type = "spearman")When we run the code above, you can see that Hmisc::rcorr returns a list. From this list, we can extract the correlation coefficients and their corresponding p-values. You can extract this list to a matrix using the extract2 function from the magrittr package or an alternative method if you prefer. We can also round each value to 2 digits to make the output cleaner.
cor.matrix.r <-
corres %>%
extract2("r") %>%
round(., 2)
cor.matrix.r mpg cyl disp hp drat wt qsec vs am gear carb
mpg 1.00 -0.91 -0.91 -0.89 0.65 -0.89 0.47 0.71 0.56 0.54 -0.66
cyl -0.91 1.00 0.93 0.90 -0.68 0.86 -0.57 -0.81 -0.52 -0.56 0.58
disp -0.91 0.93 1.00 0.85 -0.68 0.90 -0.46 -0.72 -0.62 -0.59 0.54
hp -0.89 0.90 0.85 1.00 -0.52 0.77 -0.67 -0.75 -0.36 -0.33 0.73
drat 0.65 -0.68 -0.68 -0.52 1.00 -0.75 0.09 0.45 0.69 0.74 -0.13
wt -0.89 0.86 0.90 0.77 -0.75 1.00 -0.23 -0.59 -0.74 -0.68 0.50
qsec 0.47 -0.57 -0.46 -0.67 0.09 -0.23 1.00 0.79 -0.20 -0.15 -0.66
vs 0.71 -0.81 -0.72 -0.75 0.45 -0.59 0.79 1.00 0.17 0.28 -0.63
am 0.56 -0.52 -0.62 -0.36 0.69 -0.74 -0.20 0.17 1.00 0.81 -0.06
gear 0.54 -0.56 -0.59 -0.33 0.74 -0.68 -0.15 0.28 0.81 1.00 0.11
carb -0.66 0.58 0.54 0.73 -0.13 0.50 -0.66 -0.63 -0.06 0.11 1.00cor.matrix.p <-
corres %>%
extract2("P") %>%
round(., 2)
cor.matrix.p mpg cyl disp hp drat wt qsec vs am gear carb
mpg NA 0 0.00 0.00 0.00 0.00 0.01 0.00 0.00 0.00 0.00
cyl 0.00 NA 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
disp 0.00 0 NA 0.00 0.00 0.00 0.01 0.00 0.00 0.00 0.00
hp 0.00 0 0.00 NA 0.00 0.00 0.00 0.00 0.04 0.06 0.00
drat 0.00 0 0.00 0.00 NA 0.00 0.62 0.01 0.00 0.00 0.49
wt 0.00 0 0.00 0.00 0.00 NA 0.21 0.00 0.00 0.00 0.00
qsec 0.01 0 0.01 0.00 0.62 0.21 NA 0.00 0.26 0.42 0.00
vs 0.00 0 0.00 0.00 0.01 0.00 0.00 NA 0.36 0.12 0.00
am 0.00 0 0.00 0.04 0.00 0.00 0.26 0.36 NA 0.00 0.73
gear 0.00 0 0.00 0.06 0.00 0.00 0.42 0.12 0.00 NA 0.53
carb 0.00 0 0.00 0.00 0.49 0.00 0.00 0.00 0.73 0.53 NAData Processing
Now you have the correlations and the p-values for all the variables. However, as you can see from the correlation matrix, you have repeat values for each variable with diagonal 1.000 values for correlations between the same variables. When viewing the correlation matrix, it is not helpful to see these repeat values so we can set them to missing and exclude them from the visualization using the lower.tri function from base R.
cor.matrix.r[lower.tri(cor.matrix.r)] <- NA
cor.matrix.r mpg cyl disp hp drat wt qsec vs am gear carb
mpg 1 -0.91 -0.91 -0.89 0.65 -0.89 0.47 0.71 0.56 0.54 -0.66
cyl NA 1.00 0.93 0.90 -0.68 0.86 -0.57 -0.81 -0.52 -0.56 0.58
disp NA NA 1.00 0.85 -0.68 0.90 -0.46 -0.72 -0.62 -0.59 0.54
hp NA NA NA 1.00 -0.52 0.77 -0.67 -0.75 -0.36 -0.33 0.73
drat NA NA NA NA 1.00 -0.75 0.09 0.45 0.69 0.74 -0.13
wt NA NA NA NA NA 1.00 -0.23 -0.59 -0.74 -0.68 0.50
qsec NA NA NA NA NA NA 1.00 0.79 -0.20 -0.15 -0.66
vs NA NA NA NA NA NA NA 1.00 0.17 0.28 -0.63
am NA NA NA NA NA NA NA NA 1.00 0.81 -0.06
gear NA NA NA NA NA NA NA NA NA 1.00 0.11
carb NA NA NA NA NA NA NA NA NA NA 1.00cor.matrix.p[lower.tri(cor.matrix.p)] <- NA
cor.matrix.p mpg cyl disp hp drat wt qsec vs am gear carb
mpg NA 0 0 0 0 0 0.01 0.00 0.00 0.00 0.00
cyl NA NA 0 0 0 0 0.00 0.00 0.00 0.00 0.00
disp NA NA NA 0 0 0 0.01 0.00 0.00 0.00 0.00
hp NA NA NA NA 0 0 0.00 0.00 0.04 0.06 0.00
drat NA NA NA NA NA 0 0.62 0.01 0.00 0.00 0.49
wt NA NA NA NA NA NA 0.21 0.00 0.00 0.00 0.00
qsec NA NA NA NA NA NA NA 0.00 0.26 0.42 0.00
vs NA NA NA NA NA NA NA NA 0.36 0.12 0.00
am NA NA NA NA NA NA NA NA NA 0.00 0.73
gear NA NA NA NA NA NA NA NA NA NA 0.53
carb NA NA NA NA NA NA NA NA NA NA NAIt is almost time to visualize the data. We have 2 final steps to complete the data processing. First, let’s rename the variables to something more meaningful. This way when others see them on the plot, it will be easier to understand. We will need to rename the columns with colnames and rows with rownames. We can also reshape the data using melt from the reshape2 package. Adding the na.rm = TRUE argument removes all the duplicate correlation values that were set to missing earlier.
var.names <- c("Miles/Gallon",
"Number of Cylinders",
"Displacement",
"Gross Horsepower",
"Rear Axle Ratio",
"Weight (lb/1000)",
"Quarter Mile Time",
"Straight Engine",
"Manual Transmission",
"Number of Forward Gears",
"Number of Carburetors")
colnames(cor.matrix.r) <- c(var.names)
rownames(cor.matrix.r) <- c(var.names)
colnames(cor.matrix.p) <- c(var.names)
rownames(cor.matrix.p) <- c(var.names)
cor.matrix.r <-
cor.matrix.r %>%
reshape2::melt(., na.rm = TRUE)
head(cor.matrix.r, 10) Var1 Var2 value
1 Miles/Gallon Miles/Gallon 1.00
12 Miles/Gallon Number of Cylinders -0.91
13 Number of Cylinders Number of Cylinders 1.00
23 Miles/Gallon Displacement -0.91
24 Number of Cylinders Displacement 0.93
25 Displacement Displacement 1.00
34 Miles/Gallon Gross Horsepower -0.89
35 Number of Cylinders Gross Horsepower 0.90
36 Displacement Gross Horsepower 0.85
37 Gross Horsepower Gross Horsepower 1.00The Plot
Now all we need to do is to write the code to visualize the data. Here I will use geom_tile which creates a box for each value and fills it with color based on the value variable from the data set. We can use scale_fill_gradient2 function to set the low, mid, and high colors for the correlation plot. The low and high values would correspond to -1 and 1, respectively. We can also rotate the x-axis labels by 45 degrees to make them easier to read and add the correlation values to the plot using geom_text. All of the other lines of code are to adjust the theme of the plot and can be changed to your preferences.
ggplot(cor.matrix.r, aes(x = Var2, y = Var1, fill=value)) +
geom_tile(color="black") +
scale_fill_gradient2(low="#B2BDED", mid="#ffffff", high="#C6E0B4",
midpoint=0, limit=c(-1,1), space="Lab",
name="Spearman\nCorrleation") +
theme_minimal() +
coord_fixed() +
geom_text(aes(Var2, Var1, label = value, fontface = "bold"),
color = "black", size = 4) +
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 12, hjust = 1,
face = "bold"),
axis.text.y = element_text(size = 12, face = "bold"),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
panel.grid.major = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
axis.ticks = element_blank(),
legend.justification = c(1, 0),
legend.position = c(0.5, 0.7),
legend.direction = "horizontal") +
guides(fill = guide_colorbar(barwidth = 7, barheight = 1,
title.position = "top",
title.hjust = 0.5))Warning: A numeric `legend.position` argument in `theme()` was deprecated in ggplot2
3.5.0.
ℹ Please use the `legend.position.inside` argument of `theme()` instead.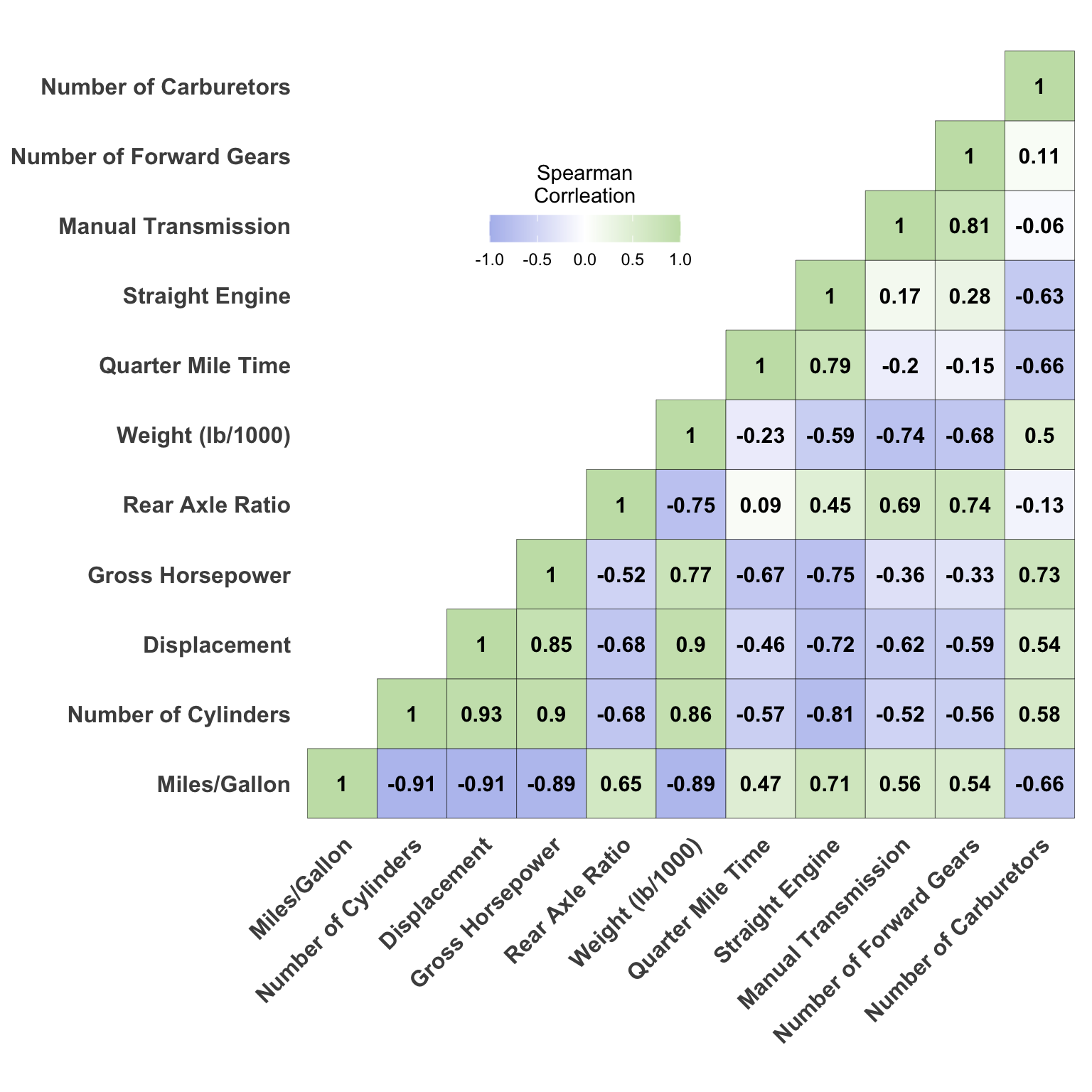
You can also add the p-values to a Table. Here I only display p-values < 0.05 if Var1 is Miles/Gallon, but you could display all p-values or adjust the filters as needed.
cor.matrix.p %>%
reshape2::melt(., na.rm = TRUE) %>%
filter(value < 0.05 & Var1 == "Miles/Gallon") %>%
mutate(value = ifelse(value < 0.001, "< 0.001", value)) %>%
gt::gt() %>%
gtExtras::gt_theme_538()| Var1 | Var2 | value |
|---|---|---|
| Miles/Gallon | Number of Cylinders | < 0.001 |
| Miles/Gallon | Displacement | < 0.001 |
| Miles/Gallon | Gross Horsepower | < 0.001 |
| Miles/Gallon | Rear Axle Ratio | < 0.001 |
| Miles/Gallon | Weight (lb/1000) | < 0.001 |
| Miles/Gallon | Quarter Mile Time | 0.01 |
| Miles/Gallon | Straight Engine | < 0.001 |
| Miles/Gallon | Manual Transmission | < 0.001 |
| Miles/Gallon | Number of Forward Gears | < 0.001 |
| Miles/Gallon | Number of Carburetors | < 0.001 |
The Function
It can be all combined into a single function. You can view the function in the visualization.R file my personal GitHub page or download it into your RStudio by typing the following code into your console.
devtools::install_github("bhelsel/bhelselR")
That’s it! Fairly simple and straightforward. There are also a lot of different ways that you could modify this function to personalize it for your analyses. Let me know if you use this function or a similar one in the comments below!